Usage
HMMpTM is freely available through http://bioinformatics.biol.uoa.gr/HMMpTM by pressing the RUN button.
Input
The submission is completed in three steps (Figure 1).
Step 1:
The input sequence must be in FASTA format. For utilizing predictions for large scale datasets users are encouraged to contact gtsaousis
![]() biol.uoa.gr
biol.uoa.gr
Step 2: The user can choose a certain score threshold for phosphorylation and glycosylation site prediction results. The default option returns all results.
Step 3: The prediction is performed by pressing the Submit button.
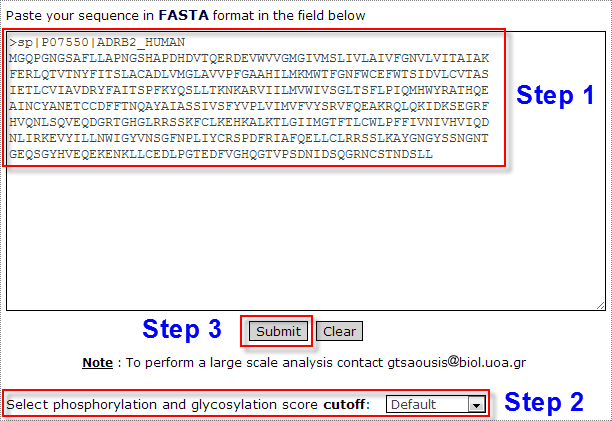
Figure 1. The input page for running HMMpTM. The submission is completed through three steps as described above.
Output
Prediction results of HMMpTM are divided into two main sections: Topology prediction results (Figure 2) and Phosphorylation and Glycosylation site prediction results (Figure 3).
Topology prediction results
In topology prediction results the user can see the protein name or the submission name as provided from the header line of the FASTA format. On the left part (Figure 2), the protein sequence along with the predicted topology is provided. The topology of the protein is provided using different symbols for predicted localizations. Possible localizations of a residue can be:
Inside ( i )
Outside ( o )
Membrane ( M )
In addition, phosphorylated and glycosylated residues with the highest posterior probability are shown using the following symbols:
Phosphorylated Serine or Threonine residues by PKA ( a )
Phosphorylated Serine or Threonine residues by PKC ( c )
Phosphorylated Serine or Threonine residues by CAMKII ( d )
Phosphorylated Serine or Threonine residues by CK1 ( e )
Phosphorylated Serine or Threonine residues by CK2 ( f )
Phosphorylated Serine or Threonine residues by MAPKs ( p )
Phosphorylated Serine or Threonine residues by CDC2 ( q )
Phosphorylated Serine or Threonine residues by GRKs ( r )
Phosphorylated Tyrosine residues by SRC ( p )
N-linked glycosylated Asparagine residues ( g )
O-linked glycosylated Serine or Threonine residues ( x )
Note that phosphorylated and glycosylated residues provided at this section are only the ones with the highest posterior probability. A more detailed list is provided at the Phosphorylation and glycosylation prediction results section.
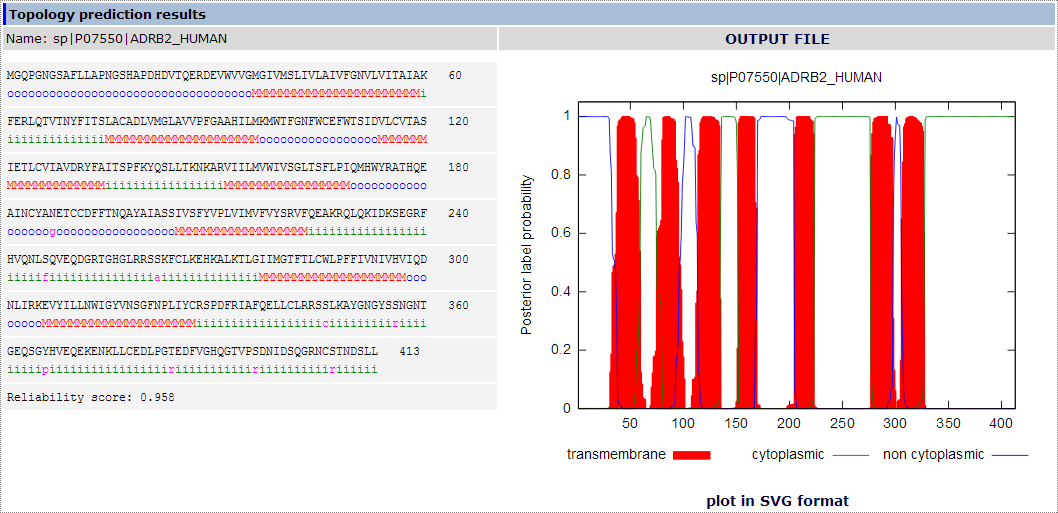
Figure 2. The topology prediction results section using the Beta-2 adrenergic receptor from Homo sapiens as an example.
Moreover, a reliability score is included for each predicted topology.
On the right part, is the posterior probability plot showing the predicted transmembrane segments in red, and the predicted cytoplasmic and extracellular loops in green and blue respectively. The plot is generated using Gnuplot (http://www.gnuplot.info/ ) and can be downloaded as a .SVG file.
Note that signal peptide prediction is not included in the HMMpTM algorithm at this point. Therefore, users should check for the existence of a signal peptide through the SignalP tool available at http://www.cbs.dtu.dk/services/SignalP/.
Phosphorylation and glycosylation site prediction results
In the phosphorylation and glycosylation prediction results section (Figure 3) we provide the amino acid residue which is predicted to be modified, its
exact location along the protein sequence and a score calculated using the posterior probability provided by HMMpTM. In the case of phosphorylation site
prediction results, the kinase responsible for the predicted phosphorylation site is provided.
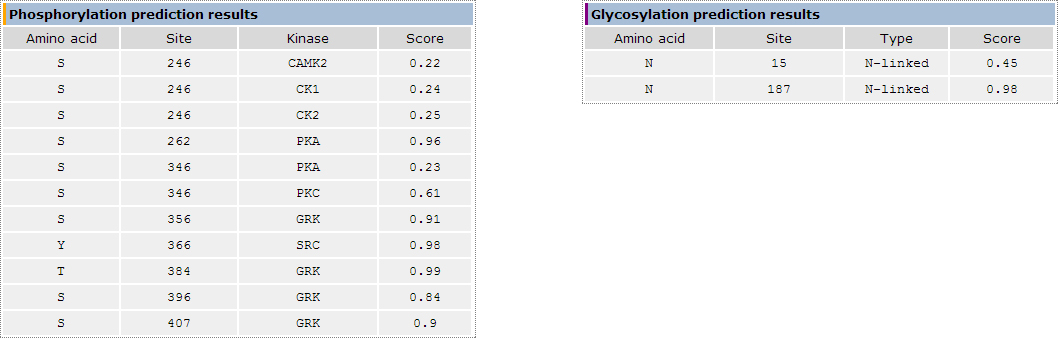
Figure 3. The phosphorylation and glycosylation site prediction results section using the Beta-2 adrenergic receptor from Homo sapiens as an example.
Download results
The user can download HMMpTM prediction results in a text file as shown in Figure 4 by pressing the "OUTPUT FILE" button on the top of the results page.
In the text version of the results the user can get the submitted protein sequence, the predicted topology and the phosphorylation and glycosylation site prediction results with extensive information.

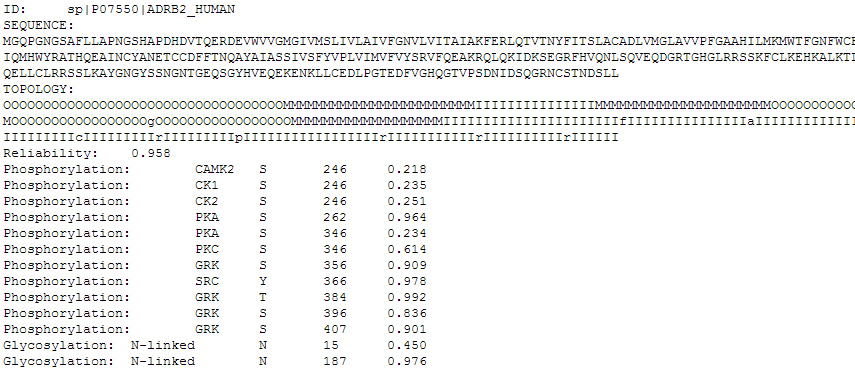
Figure 4. The text output file of HMMpTM. Here is the example of the Beta-2 adrenergic receptor from Homo sapiens.
Note that the text output file is maintained in the server for 72 hours and can be accessed through the provided link (using a unique serial number for each submission, e.g. 1368636225). All submissions and results are kept confidential and deleted after 72 hours.
Below is an example of the provided link: http://aias.biol.uoa.gr/HMMpTM/TMP_FILES/1368636225.hmmptm.txt
Datasets
All datasets used in HMMpTM are available for download through here

