
DETECTION OF THE FOUR DISTINCT Galpha FAMILIES, THE Gbeta AND THE Ggamma SUBUNITS USING profile Hidden Markov Models (pHMMs)
Instructions
Heterotrimeric G-proteins are molecular switches that turn on intracellular signaling cascades in response to the activation of G-protein coupled receptors (GPCRs) by extracellular stimuli in eukaryotes. They are composed of three subunits, α, β and γ. Their nomenclature is determined by their α-subunit and they can be classified in four families in mammalians: Gs, Gi/o, Gq/11,G12/13.Our goal is to detect and classify members of the four distinct families, plus the Gβ and the Gγ subunits of G-proteins from sequence alone
Here, we present a profile Hidden Markov model method that detects members of the four distinct families, the Gβ and the Gγ subunits of G-proteins from sequence alone. For this purpose we have constructed six specific profile Hidden Markov Models (pHMMs) for each Gα subunit family, as well as for Gβ and Gγ subunits. In order to construct these pHMMs, we implemented multiple alignments, which then were used as input in the HMMER package (Eddy, 2006; Eddy, 1998). In order to build the pHMMs we used not only positive but also negative training sequences so as to make the models more specific (HMM-ModE) (Srivastava, et al., 2007). After the build process, the six pHMMs were converted to HMMER v3.0. The six models that were developed were then tested for their reliability against the whole UniProt/Swiss-Prot database. We finally defined the cutoff value as: (TC+NC)/2, where TC stands for Trusted Cutoff and NC as Noise Cutoff.
The following table shows the cutoffs for the six profiles that we have constructed:
| Gs | Gi/o | Gq/11 | G12/13 | Gbeta | Ggamma | |
|---|---|---|---|---|---|---|
| Cut-off | 608.3 | 770.7 | 735.3 | 632.2 | 291.5 | 34.4 |
Moreover, our tool incorporates an additional pHMM from Pfam database (id: PF00503) that describes the G-protein alpha subunit in general, without further distinction among the four known G-protein alpha families.
You can use our method as below:
- Copy and paste your sequence(s) in FASTA format into the submission form or upload a file.
- Press “Submit Query”.
- You can also retrieve again your results by inserting your Job ID. The results will be available at least for 24 hours.
A results page will then be returned :
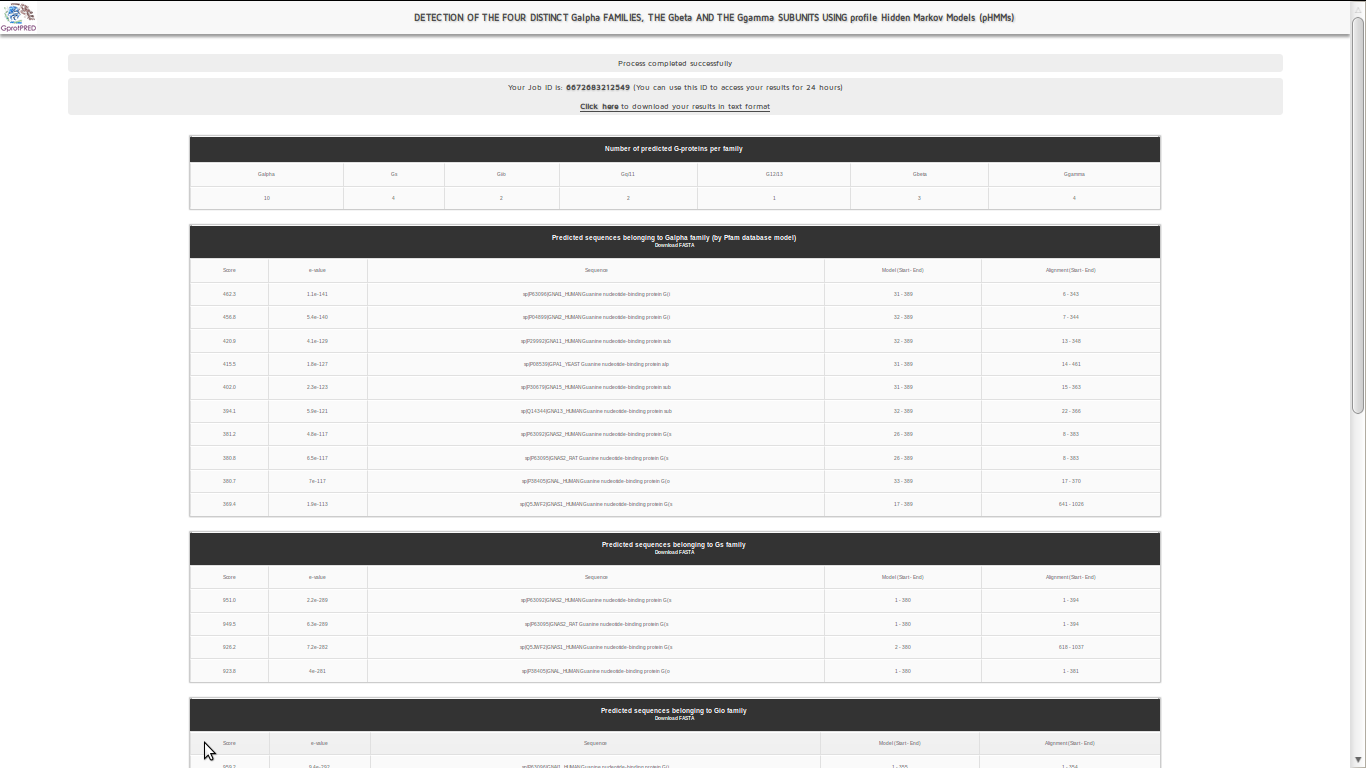
Seven different result tables containing the predicted sequences for each pHMM separately will then appear as well as your unique Job ID. Each table shows the predicted G-proteins in accordance with the user’s data and contains HMMER score(s), e-value(s), the sequence(s) description as well as the regions that are aligned in the model and the sequence respectively. There will be also an extra table which summarizes the total number of predicted G-proteins according each profile.The user will also be able to download the FASTA files of the predicted proteins by clicking the link Download FASTA. A result file in text format is also available.